
Magnetic resonance tools, easy to use.

Magnetic resonance tools, easy to use.
DNPy is a Python code package to evaluate continuous wave (CW) Overhauser dynamic nuclear polarization (ODNP) experiments. DNPy has taken the core functionality from pyNMR.
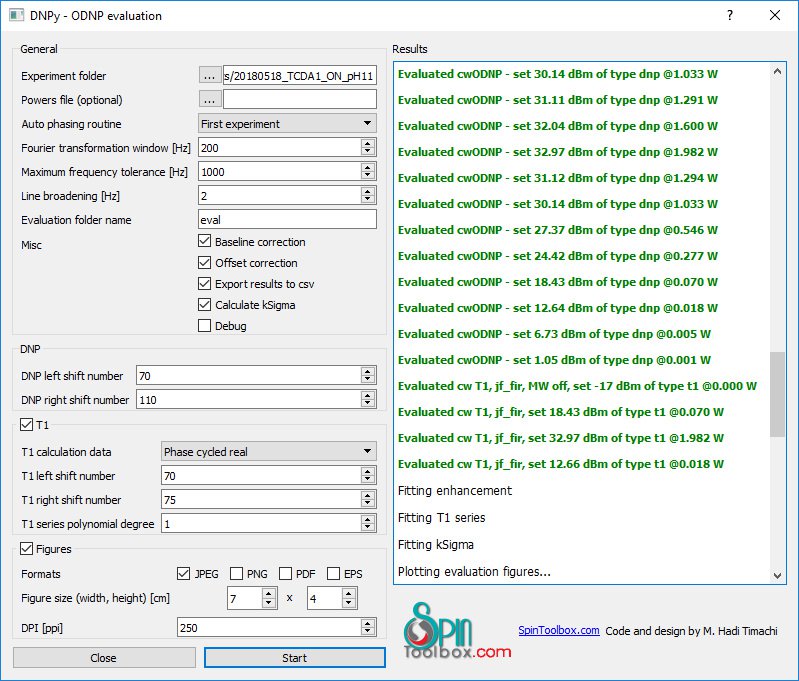
To be able to use this code package, you need to have power set written in your experiment title with the following style:
XXXXX - set XX dBm example: CWODNP - set 10 dBm, T1 experiment - set 5 dBm
If you have a calibration curve based on main attenuation setting, you can use the following instead:
XXXXX - set XX dB example: CWODNP - set 10 dB
This however requires you to have a data calibration file as a CSV when using DNPy which should have comma as a separator and 2 columns (dB set - dBm output).
You need to have python3 with following packages installed:
pip3 install --user numpy or sudo apt install python3-numpy on linux.pip3 install --user scipy or sudo apt install python3-scipy on linux.pip3 install --user matplotlib or sudo apt install python3-matplotlib on linux.pip3 install --user pyqt5.Additionally, if you want to have JPG output plots, you need to install pillow: pip3 install --user pillow.
You can install ANCONDA for windows to have all necessary packages installed (don't forget to pick python3 package). After installation you can run the folowing in the command prompt: C:\ProgramData\Anaconda3\python.exe DNPyUI.py. Installation directory can differ based on your selection when installing Anaconda.
DNPyUI can be run with the following. It will show you an intuitive UI.
python3 DNPyUI.py
If you prefer to use DNPy as a function call, you can import functions and setup kwargs for return_exps() function. A sample usage is implemented in dnpEval.py. You need to set the following variables in your dnpEval:
path = '/path/to/exp/folder/' powerFile = 'powers' kwargs = { 't1Calc': 'PCreal', # 'PCreal' for real phase cycled channel or 'PCmagn' or 'real' or 'magn' 'phase': 'first', # all, none, first 'ftWindow': 200, # X domain for FT plot, int. and phase calculation 'maxWin': 1000, # Does not allow peaks out of this domain to be calculated in int. 'lS': 70, # Left shift points or 'auto' 'lSt1': 70, # LS for t1 experiments (this differs from DNP sometimes) 'rS': 110, # Right shift points 'rSt1': 75, 'lB': 2, # Line boradening [Hz] 'offCor': True, # offset correctioon 'basCor': True, # baseline correction 'evalPath': 'evalHadiTest', 'plotDpi': 250, # plot file resolution 'plotExts': [], # remove all if you do not want plots to be saved 'process': True, 'debug': False, 'dumpToCsv': True, 'figSize': (13, 8), 'powerFile': powerFile, 't1SeriesEval': True, 't1SeriesPolDeg': 1, # Polynomial degree for T1 series fit (default = 1) 'kSigmaCalc': True, }
path defines the ODNP experiment root folder.powerFile is used when you have "ODNP - set .. dB" style title in your experiments and is a csv file containing dB to dBm conversion table.debug boolean turns verbose evaluation on and gives you more information.plotExts denotes the plot file extensions you want to have saved.Finally, we call the main function using:
exps = return_exps(path, **kwargs)
DNPy can be downloaded from the github page.
Created: 2018-06-07 Thu 09:44
Modified: 2018-07-02 Mon 15:56
2018.06.29:
2018.06.07 initial release: